Result and Discussion
Eight strains (CS1 e CS8) exhibited positive characteristics to extracellular lipase secretion on agar medium based
lipase screening tests among the twenty-four fungal strains (endophytic/ fee-spores) isolated from the high saline
region,. These eight strains were further scaled down to one strain (CS4) based on the obser- vation of
distinctively higher zone of clearance, usually measured by the degradation halo of enzyme-specific substrates.
Morpho- logical examination of conidia and conidiophores under high- resolution optical microscope after lactophenol
cotton blue stain- ing revealed the pattern of conidial spores belonging to the genera Cladosporium sp. SEM
visualization of the grown hyphae and mycelia reconfirmed the identification of filamentous fungus to be Cladosporium sp.
The light microscopy and SEM images of the Cladosporium sp. is presented in Fig.
3. The results of tween80 - CaCl2 and olive oil - phenol red-based lipase screening test
images
Fig. 3. Light microscopy and SEM images of Cladosporium sp. CS4.
displaying zones of clearance are presented in Fig. 4. The molecular identification and phylogenetic analysis of the strain CS4 revealed
maximum homology with the fungus Cladosporium tenuissimum, identified hereafter as Cladosporium sp.
CS4 is presented in the form of a phylogenetic tree in Fig. 5. The prospects of lipase
extracted from this genera as a potential biocatalyst for biodiesel production is reported for the first time through
this work though there are some reports of Cladosporium sp. extracted lipase [22,62]. Two different strains of Cladosporium cladosporioides were earlier [62] isolated and reported from rotten maize for deter- mining variations in lipase
enzyme activity, revealed exceedingly higher levels of lipase activity on agar plates evident from the higher zone
of clearance among a total of twenty-three other fungal isolates. Moreover, the lipolytic activity exhibited by
C. Cladosporioides was much higher than that of P. solitum [63].
Enzyme production, purification and molecular weight determination
The ambient conditions of all enzyme purification steps were maintained at 4 C. Cladosporium sp. CS4 produced
extracellular crude lipase was subjected to partial purification/protein
concentration using ammonium sulphate precipitation. The pellet obtained after ammonium sulphate precipitation was
suspended in
0.5 M Tris-Chloride buffer at pH 7.2 and dialyzed using 3.5 KDa Molecular Weight Cut off (MWCO) against the same
buffer with
0.1 M NaCl. Gel filtration chromatography was performed by elution of partially purified enzyme using buffers
containing appropriate salt concentrations after proper degassing of buffers and deionized water used for
equilibrating the column. Gel filtration-based size-exclusion chromatographic system uses a hy- drophilic packing
material and an aqueous mobile phase to frac- tionate water soluble proteins based on their differences in its
molecular weight. When dissolved protein molecules of various sizes flows through the column, larger proteins elute
first as the pores of the hydrophilic gel packed inside the column is too small for the larger proteins to enter,
whereas smaller proteins elute slower after entering the pores of the packed gel column thus leading to effective
sorting and separation of proteins based on size [64]. Superdex 200 column filtration of
the enzyme resulted in a purification fold of 4.1 with an activity recovery (yield) of about 36.7% and specific
activity of about 37.2 U/mg. Table 1 lists the gradation in improved activity, purity and
yield at each stage of enzyme purification. The present study show comparable yield
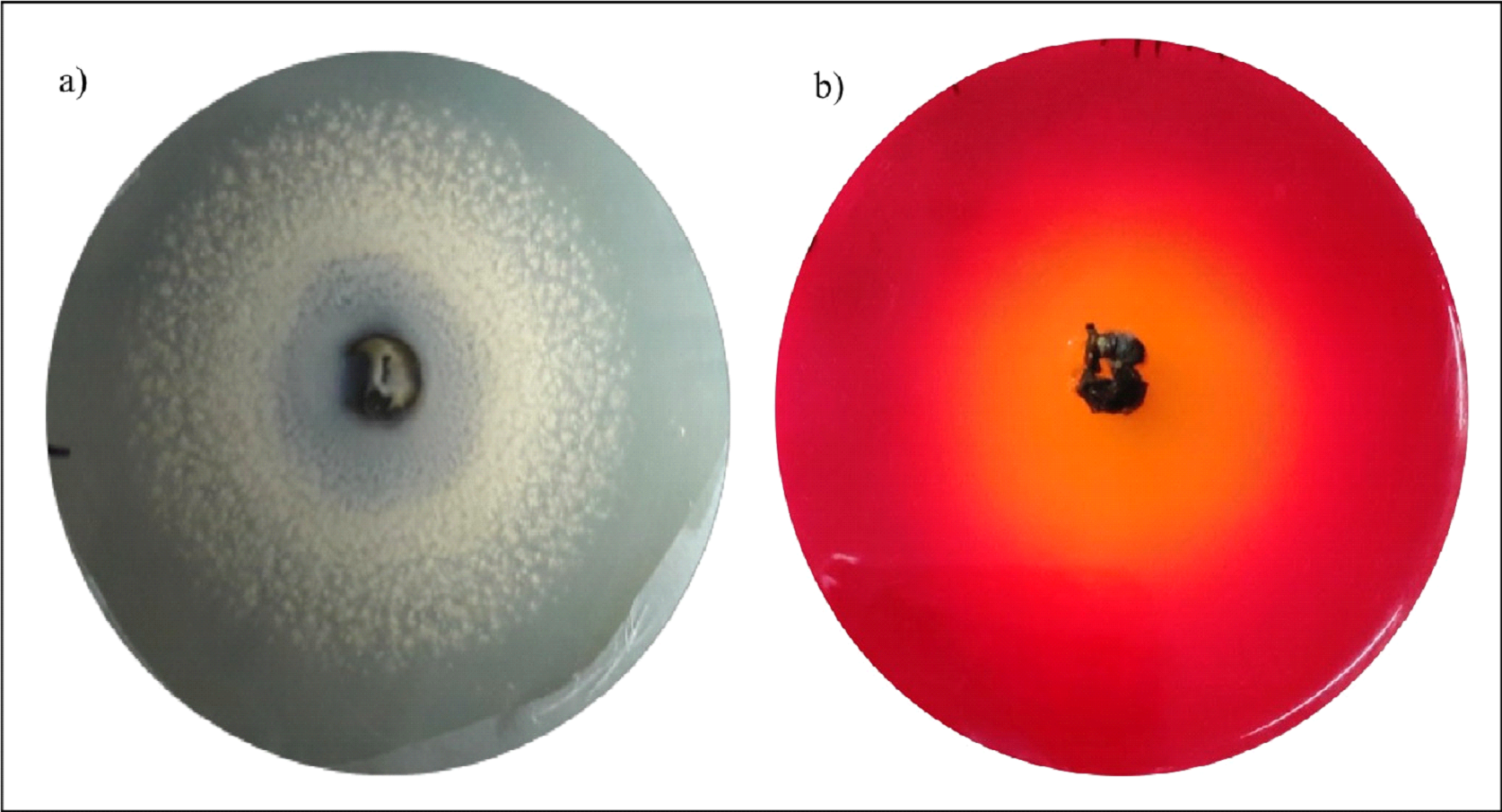
Fig. 4. Degradation halo zones exhibited by Cladosporium sp. CS4 in a) CaCl2-tween 80 and b) phenol red-olive oil agar plates.
Fig. 5. Phylogenetic analysis exhibiting homology of strain CS4 with Cladosporium tenuissimum
images
Table 1
Summary indicating improvement in purification after each enzyme purification step.
Diatom |
Lipid Content (%) dcw |
Reference |
Achnanthes delicatulum hauckiana |
29.8 |
[105] |
Achnanthes sp. |
27.7 |
[106,107] |
Aulacoseira Ambigua |
19.7 |
[108] |
Bacillaria paradoxa |
33.61 |
[109,110] |
Cocconeis peltoides |
20.9 |
[105] |
Chaetoceros curvisetus |
14.86 |
[111] |
Diatoma sp. |
15.76 |
[105,112] |
Cocconeis sp. |
31.81 |
[113] |
Cyclotella cryptica |
27 |
[114] |
Melosira sp. |
14.75 |
[115] |
Nitzschia dissipata var. media |
37.5 |
[114,116] |
Phaeodactylum tricornutum |
10.7 |
[117] |
Synedra ulna |
7.58 |
[55] |
Tryblionella navicularis |
24.2 |
[105] |
Nitzschia punctata |
16 |
This study |
with higher purification than reported earlier [33]. Lipase extrac- tion and
characterization on a yeast strain Ralstonia sp. resulted in 3.9-fold purity and 20.8% yield respectively [33]. Lipase purification on a biocontrol fungus Nomuraea rileyi exhibited a
yield of 1.69% with a purification fold of 23.9 [65]. Fungi Penicillium expansum had
reported the highest purification fold of 219.0, with a poor yield of 5% [66]. Purification
and characterization of lipase extracted from a bacterium Pseudomonas aeruginosa reported 8.6-fold
purification with 51.6% activity recovery [51]. After purification, aliquots of 40 mL crude
culture filtrate, partially purified and purified protein was loaded onto SDS PAGE with 10% polyacrylamide gel to
observe visible bands of lipase as against protein molecular markers. The molecular weight of the purified protein
was ~46 KDa through SDS PAGE. SDS PAGE gel representing different lanes with enzyme and protein molecular marker is
given in Fig. 6.
Enzyme characterization
Effects of pH and temperature
The structural and functional relationships between enzymes and reaction media are primarily determined by ambient
envi- ronmental conditions, especially pH and temperature [67]. Fig.
7 represents varying temperature and pH versus relative and resid- ual enzyme activity. The lipase activity
of the enzyme was maximum at pH 6 and at a temperature of 60 C. The relative enzyme activities at pH 4.0 and pH 5.0
were 34 ± 7% and 31 ± 8% to that of the maximal lipase activity observed at pH 6.0. Significantly higher lipase
activity was recorded at pH 6.0, with <35% of the relative activities observed at other pH ranges depicting the
sensitiveness of lipase to varying pH ranges. Mycelium bound intracellular lipase extracted from the fungus Aspergillus
niger showed optimal pH in the acidic range from 3.0 to 6.5 with higher activity at the pH of 4.0 [68]. Candida cylindracea yeast extracted lipase [69] showed maximum lipase activity at pH 6.0. However, intracellular lipase
extracted from fungi Aspergillus westerdijkiae [70], Nomuraea
rileyi [42] and Rhizopus oryzae [71]
exhibited optimal pH activity on the alkaline pH (8.0e8.5). The optimum temperature at which the lipase enzyme
showed maximum activ- ity was 60 C and fairly high stabilities were recorded at higher temperatures of 40 C (95 ±
1%) and 50 C (99 ± 1.5%), which are comparable to the performance of Mrakia blollopis yeast extracted
lipase [49].
Effect of surfactants on lipase activity
Long-chain ionic liquids like surfactants have a tendency to alter its physicochemical properties by internal
alteration of charges
Fig. 6. Characterization of enzyme at each stages of purification.
from cationic to anionic and vice versa [72]. These surfactants aid in providing enzymes
with access to substrates through interfacial area stabilization, which enhanced the catalytic reaction of lipase
[50]. Surfactants are important components of emulsion prepara- tions during lipase
assays at every stage from enzyme production, purification and characterization [73]. Fig. 8 illustrates the effect of different surfactants on lipase enzyme activity.
Lipase enzyme showed variation in affinities to each substrate, when introduced into different concentrations of
surfactants like Triton X100, SDS, Tween 80 and Tween 20. Triton X100 had less affinity to enzymes with least
activity exhibited among other surfactants. The addition of varying percentage concentrations of SDS increased the
lipase activity with maximum activity recorded at SDS concentration of 0.25% with 4.02 ± 0.27 U/mL. Higher
concentrations of SDS was found to inhibit the enzyme activity. Enzyme activity was less yet uniform among different
concentration ranges of tween 80, how- ever, for tween 20, activity for a concentration of 0.5% (2.89 ± 0.61 U/mL)
was found to be the maximum. Fungus Nomuraea rileyi showed enhanced lipolytic activities in the presence of
SDS and tween 80 which is similar to the results of the present study [73], whereas
lipase extracted from thermophilic Rhizopus oryzae and a yeast Yarrowia lipolytica showed
decreased with inhibited activities when introduced to SDS and tween 80 [74,75].
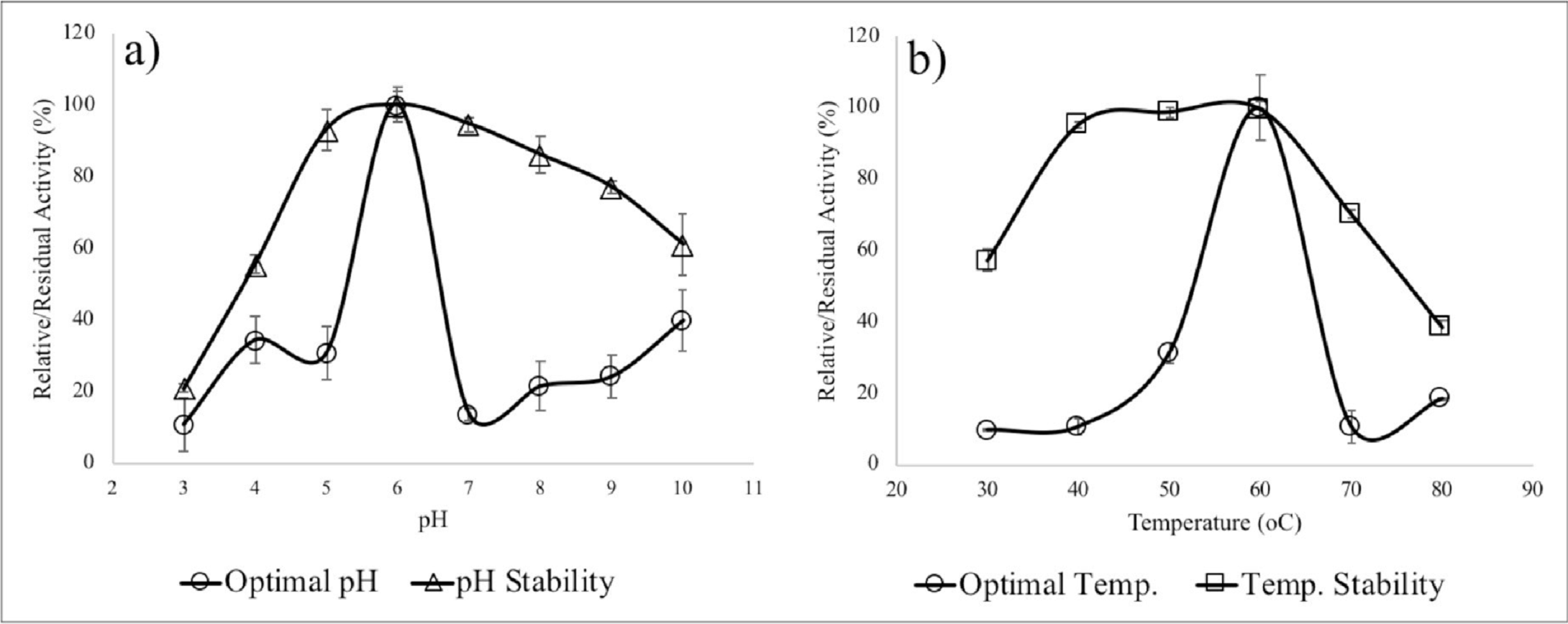
Fig. 7. a) Effect of pH on lipase activity and pH stability of lipase b) Effect of
temperature of lipase activity and residual activity at different temperatures.
Fig. 8. Effect of different surfactants and its concentration on lipase enzyme activity.
Isolation, growth and SEM analysis of microalgae
Initial agar plating (spread plate method) and subculturing by quadrant streaking of epipelic diatoms (diatoms
attached to sedi- ments) collected from the salt pan, resulted in the isolation of pure diatoms. Morphological
characterization of the diatom isolates using high-resolution phase contrast optical microscope and SEM
visualization aided in species-level identification of the diatom. Comparison of high-resolution images with standard
identification keys confirmed the diatom as Nitzschia punctata. It is a pennate diatom with valves
elliptic-lanceolate and apices rostrate, striae more or less curved, with transverse, radiant and moniliform forming
coarse puncta placed at equal distances [53,76]. Apices have
no nodules with often showing fine scattered puncta. The alga measures ~13. 5e14. 5 mm in length and ~5.6e6.0 mm in
width. The light microscopy images of Nitzschia sp. in a colony with evident extracellular polymeric
secretion (EPS) matrix in the background
and SEM images of the culture is presented in Fig. 9.
Algal biomass concentration and productivity determination
Diatoms grown in 10-L culture tubs at the rooftop in the natural daylight were harvested after nine days of
cultivation period. The algal biomass was harvested through simple siphoning of spent culture medium and washed
thrice using deionized water to ensure the complete removal of traces of salt from algal biomass and subsequently,
oven-dried at 85 C overnight. The dried diatom
biomass exhibited a biomass concentration of 354 ± 0.24 mg L-1 and biomass productivity of 39.3 ± 0.14 mg
L-1 d-1. Benthic diatom Navicula cincta [77] reported a
biomass concentration of
318.5 mg L-1 [78] and diatom Cylindrotheca closterium resulted in
biomass production of 356 mg L-1. Similar work [79] that compared
growth rate and biomass productivities of twenty-two different genus of microalgae reported the biomass
concentrations and
Fig. 9. a, b) Light microscope images of live
Nitzschia punctata culture with a
matrix of EPS at the background c, d) SEM images of processed
Nitzschia culture f) acid digested silica
frustules of
Nitzschia sp. imaged under 100x oil immersion in phase contrast optical microscope.
productivities of two different strains of Cylindrotheca fusiformis to be 357 and 366 mg L-1 and
55 and 52 mg L-1 d-1 respectively. The biomass concentration and productivity values reported
in other studies are very much comparable to the present result. Only a hand few studies that concentrate on
biodiesel prospects of di- atoms among other microalgae [55,78e82]. Nitzschia sp. was explored for its potential as a third-generation
feedstock for bio-
diesel production is reported for the first time.
Lipid extraction and quantification
In-situ environmental parameters during diatom growth act as a decisive factor in the accumulation of lipids and the
lipid content [83]. Generally, nitrate and silicate nutrient deprivation in diatoms has
induced higher lipids [84]. The quantity of lipids present in Nitzschia sp. was
gravimetrically determined after lipid extraction using Soxhlet apparatus. The culture showed 16% lipid content with
an estimated oil yield 16% of the algal biomass used for lipid extraction based on dry cell weight after nine days
of outdoor cultivation under sunlight. As the diatoms were grown under nutrient replete conditions, the lipid
content is comparatively lower than the values reported in other studies on diatoms. Table
2
Table 2
Lipid content across different diatom species.
Fig. 10. Fatty acid profile of lipids extracted from
Nitzschia sp.
lists the estimated lipid contents of different genera of a pennate diatom.
Fatty acid characterization
Fatty acid profiles of biofuel feedstock help in assessing biodiesel quality [85].
Biodiesel quality is significantly governed by the na- ture of fatty acids in terms of percentage composition, degree
of saturation/unsaturation and the length of the carbon chains pre- sent [86,87]. Fig. 10 illustrates the peaks of fatty acids
extracted from Nitzschia sp. Fatty acid profiling of lipids extracted from Nitzschia punctata resulted
in higher proportions of C16 and C18 fatty acids (Table 3). GC-MS peaks recorded the
presence of nine different fatty acids, with a predominance of saturated fatty acids. Long-chain fatty acids such as
linoleic acid, trans-vaccenic acid and arachidic acid were effectively converted into methyl esters by lipase
biocatalyst extracted from Cladosporium sp. CS4, which highlights selective and highly specific enzymatic reactions
on microalgal oil. Similar fatty acids profile was reported earlier in marine microalgae Tetraselmis sp. [88].
FAME composition analysis and biodiesel yield estimation
The compositional distribution of FAMEs is listed in Table 4. Fig.
11 highlights GC-MS peaks representing different FAMEs eluted in acid-catalyzed (ACT) and lipase-catalyzed (LCT)
treatments of diatom oil. Ten different FAME mixtures were observed in GCMS peaks of acid-catalyzed and
enzyme-catalyzed biodiesel. FAMEs showed a dominance of saturated fatty acids (SFAs) with larger proportions of
C16:0 (palmitic acid methyl esters) followed by monounsaturated fatty acid (MUFAs) C18:1 (oleic acid methyl es-
ters). An increase in saturated and monounsaturated fatty acids with an increase in salinity in Dunaliella sp.
was reported with a significant decrease in polyunsaturated fatty acids (PUFAs). methyl
Peak No. RT (min.) Fatty acids Peak area Peak area (%)
linoleic acid (C18:2) n-3 which were
absent in acid-catalyzed biodiesel sample made its presence in lipase-catalyzed biodiesel [89].
The similar appearance of methyl esters of monounsaturated fatty acids was found in NaCl treated cells of Chlorella sp.
to that of control without NaCl was reported by Ref. [90]. In enzyme- catalyzed
biodiesel, mix of SFAs and MUFAs - palmitic (53.12%), oleic (24.25%) and linoleic (11.05%) acid methyl esters
constituted up to 88.4% of the total fatty acids present in the sample, whereas in acid-catalyzed biodiesel mixes of
SFAs and PUFAs - palmitic (55.22%), myristic (5.95%) and pentadecanoic acid (3.28%) and octadecanoic acids (16.56%)
constituted up to 84.65%. Algal oil with a higher composition of PUFAs is inappropriate as fuel for engines as PUFAs
lead to lower oxidative stability of the fuel which in turn affects engine performance [91].
The qualitative analysis of Nitzschia sp. generated FAMEs through lipase-catalyzed transesterification showed
a predomi- nance of saturated (62.83%) and MUFA (37.0%), thus possessing the characteristics to produce good quality
biodiesel with higher oxidative stability. On the other hand, in acid-catalyzed biodiesel C16:0 and C18:1 were
predominant however, MUFAs palmitoleic, linoleic and trans vaccenic acid were absent. An ideal mix of C16:1, C18:1
and C14:0 in the mass ratio of 5:4:1 produce superior quality biodiesel with higher cetane number (CN), good cold
filter plug flow properties (CFPP) and better oxidative stability [92,93]. FAMEs generated by lipase-catalyzed transesterification has not yielded C16:1,
C18:1 and C14:0 at exact proportions, but GC-MS shows the presence of all three important FAMEs. Tweaking
environmental parameters would result in good quality biodiesel with prescribed fatty acid composition [92]. Biodiesel yield cal- culations were carried out on LCT and ACT samples using [13] highlight higher FAME yield of 87.24% for lipase-catalyzed trans- esterification
when compared to acid-catalyzed transesterification yield of 83.08%. Thus, FAME yield confirms the superior perfor-
mance of biocatalyst over conventional chemical catalysts in the transformation of microalgal oil into biodiesel.
FT-IR analysis of algal oil and biodiesel
FT-IR spectrum of algal oil and biodiesel derived from algal oil
by acid and enzyme-catalyzed transesterification is given in
Fig. 12aed. Fig. 12a represents the IR spectrum of hexane extracted
algal fatty acids, while Fig. 12b and c represents FAMEs extracted
from algal oil using acid (ACT) and enzyme (LCT) catalysts. Fig. 12d
represents the comparative graph of all three FT-IR peaks together.
FT-IR bands at 3009.5 cm1
, 2854.08 cm1 indicates the presence
of C e H stretching vibrations of alkanes while the peak of
1465.28 cm1 indicates bending vibrations of C e H alkane bonds
(methylene group) [94] reflected from hexane as the fatty acids
and FAMEs are extracted in hexane. The peak at 1379.5 cm1 is due
to symmetric bending of ds (CH) CH3 vibrational modes, a probable
1-hexene derivative of hexane. The weak band at 1238.3 cm1 is
due to strong stretching vibrations of (C e O) alkyl aryl ether [95].
The strong peak at 1745.44 cm1 represents C ¼ O stretching of
esters in methoxy carboxylic acids confirms the presence of higher
proportions of free fatty acids and FAMEs [96,97]. The peak at
1465.28 cm1 represents the medium C e H bending stretch of
alkane (methylene) group. The peak 1163.15 cm1 represent
stretching vibration of C e O e C ester (formates) groups. The
vibration at 722.56 cm1 is due to bending vibrations of ¼ C e H of
alkenes and aromatics [98]. Extra peaks at 1157 cm1
, 955.9 cm1
and 724 cm1 in the lipase-catalyzed biodiesel, indicates strong C
e O stretching of aliphatic ether and C ¼ C bending of alkene
compounds respectively. The FT-IR highlights the predominance
of C e H group confirming the suitability of algal oil as biodiesel
Table 4
Compositional variation of acid catalyzed and enzyme catalyzed microalgal (diatom) biodiesel.
[99], similar to the earlier studies [60,96,100] for vegetable oil as
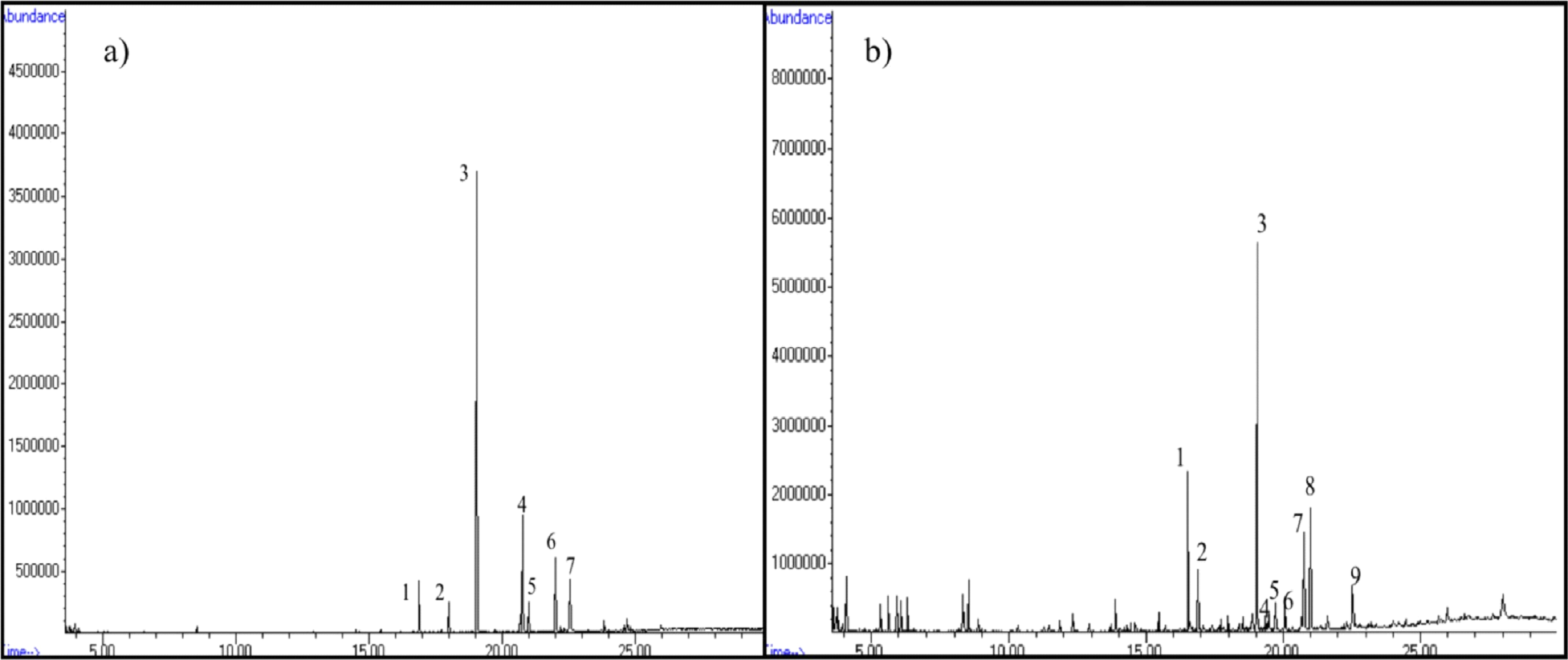
Fig. 11. FAME composition of a) ACT and
b) LCT diatom biodiesel.
well as algal oil-derived biodiesel.
The analyses confirm the prospects of using fungal lipase as biocatalyst for effective conversion of algal oil to
biodiesel. The results indicate the suitability of Cladosporium tenuissimum extracted lipase as a low-cost
biocatalyst over conventional acid catalysts with assured environmentally-friendly bioproducts. Optimization of
reaction conditions such as reaction temperature, agitation speed, the volume of methanol and enzyme for enzyme- catalyzed transesterification through design of experiments would
lead to enhanced FAME yields. Moreover, experiments utilizing Cladosporium tenuissimum as a whole-cell
biocatalyst in free or immobilized form would open new avenues of sustainable bio- diesel production through a scope
for enzyme reuse, which reduces production costs.
Comparative asseswsment of biodiesel yield
Lipase catalyzed transesterification of microalgal oil is usually
images
Fig. 12. FT eIR spectra of fatty acid and FAME extracted from Nitzschia sp.
Table 5
Comparison of FAME yields in lipase catalyzed transesterification of different generations’ biodiesel feedstocks.
Feedstock used |
Lipase extraction source (organisms) |
Type/Nature of Lipase |
FAME yield (%) |
References |
Sardine oil |
Rhizopus oryzae |
Immobilized |
76.5 |
[74] |
Sardine oil |
Mucor hiemalis |
immobilized |
38.4 |
[118] |
Sardine oil |
Candida rugosa |
immobilized |
50 |
[119] |
Soybean oil |
Pseudomonas cepacia |
Gel entrapment |
67 |
[120] |
Jatropha oil |
Chromobacterium viscosum |
Celite 545 entrapment |
71 |
[121] |
Waste oil, plant oil |
Candida sp. |
Cotton membrane immobilization |
90e92 |
[122] |
Chlorella vulgaris ESP 31 |
Burkholderia sp. C20 |
immobilized |
92.15e95.75 |
[123] |
Gracilaria edulis |
Candida antartica lipase in Pichia pastoris (Cal A, Cal B) |
Free, Immobilized |
88.5e93 |
[59] |
Enteromorpha compressa Candida antartica lipase in Pichia pastoris (Cal A, Cal B) Free,
Immobilized |
89e92 |
[59] |
Ulva lactuca Candida antartica lipase in Pichia pastoris (Cal A, Cal B) Free,
Immobilized |
87e90 |
[59] |
Scenedesmus obliquus |
Aspergillus niger |
Immobilized in BSPs |
90.82 |
[37] |
Chlorella salina |
Rhodotorula mucilaginosa |
Immobilized |
85.29 |
[34] |
Chlorella sp. |
Mold-fungus JN7 |
Immobilized in BSPs |
50.3 |
[124] |
Marine microalgae DY54 |
Mold fungus JN7 |
Immobilized in BSPs |
68.2 |
[124] |
Chlorella protothecoides |
Candida antarctica (type B) |
Free and Immobilized in combination |
97 |
[36] |
Tetraselmis sp. |
Candida rugosa |
Commercial |
7 folds higher FAME conversion rate |
[20] |
Nitzschia punctata |
Cladosporium tenuissimum |
Free |
87.2 |
This study |
carried out in two different ways: i) using lipase enzyme either extracted from extracellular or intracellular enzyme
secretions of fungi, yeast or bacteria as a source of catalyst and (ii) Using fungal/ bacterial (immobilized) cell
as a whole cell lipase. Microbacterium sp. [101], Pseudomonas sp [102,103]. are the bacterium explored for its lipase and
Aspergillus sp. [37], and Candida sp [104].
are the fungal and yeast species explored earlier. However, Cladosporium sp. extracted lipase used for the
production of microalgal biodiesel is a first of its kind study with prospective scope for future research. Earlier,
Chlorella sp. and Scenedesmus sp. were the widely studied microalgal species for lipase mediated
transesterification with re- ported FAME yields ranging between 85 and 97% [37,41,102]. The analyses of diatom lipids for the
production of biodiesel using lipase provide important insights on the potential of diatoms to be explored for
biodiesel prospects. Comparison of FAME yields in lipase catalyzed biodiesel production from available literature
with that of the current study is listed in Table 5.